An Introduction To Java Web Applications
Introduction
A Web application is an application that is accessed over a network such as the Internet or an intranet. While the earliest websites served only static web pages, dynamic response generation quickly became possible via CGI scripts, JSPs (JavaServer Pages), servlets, ASPs (Active Server Pages), server-side JavaScripts, PHP, or some other server-side technology.
Java has become a popular language for creating dynamic Web applications over the last 15 years, due to the introduction of servlets, JSP, and frameworks such as JSF and Spring. In this post we give an overview of these technologies, and explain the the major differences between them.
Building Blocks
For the sake of simplicity we distinguish three types of Web application building blocks: servlets, JSPs and frameworks
Servlets
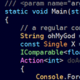
In Java, Web applications consist of servlets. A servlet is a small Java program that runs within a Web server. Servlets receive and respond to requests from Web clients, usually across HTTP. An example of a servlet that takes a request and returns a page with the numbers 1 to 10 is given below.
import java.io.*; import javax.servlet.*; import javax.servlet.http.*; public
…